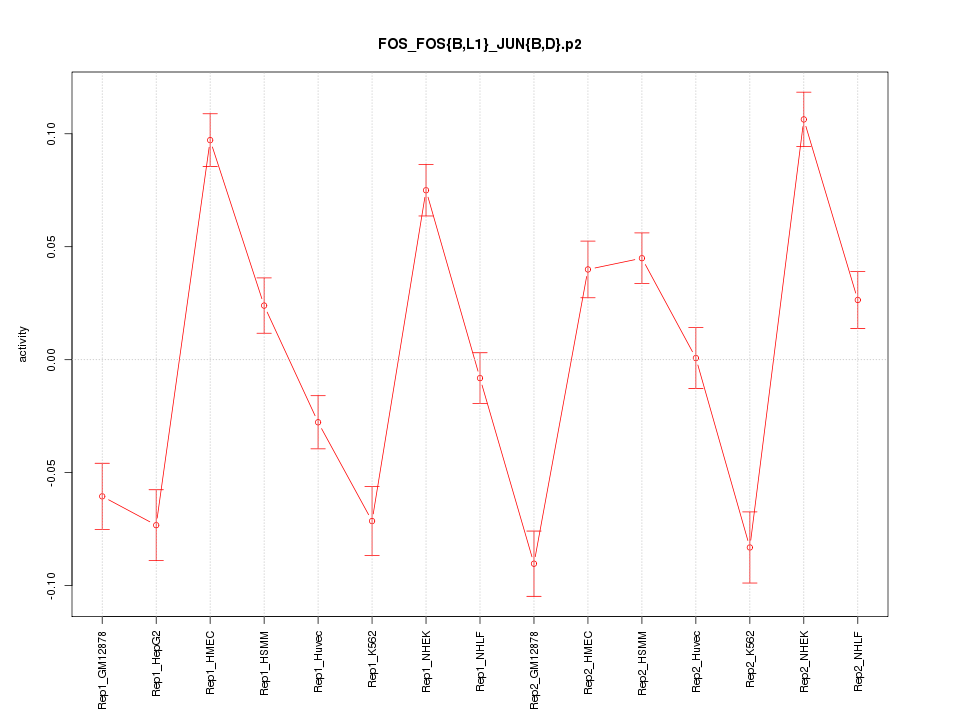
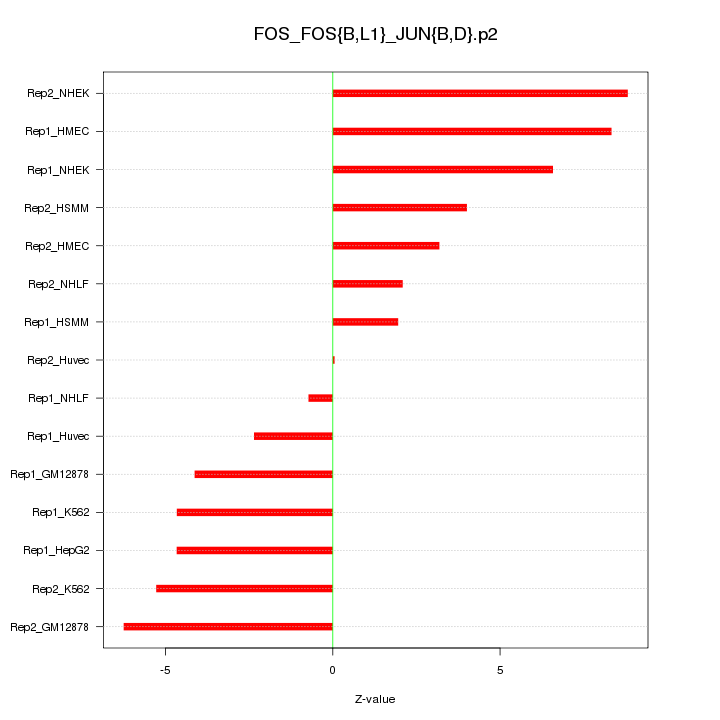
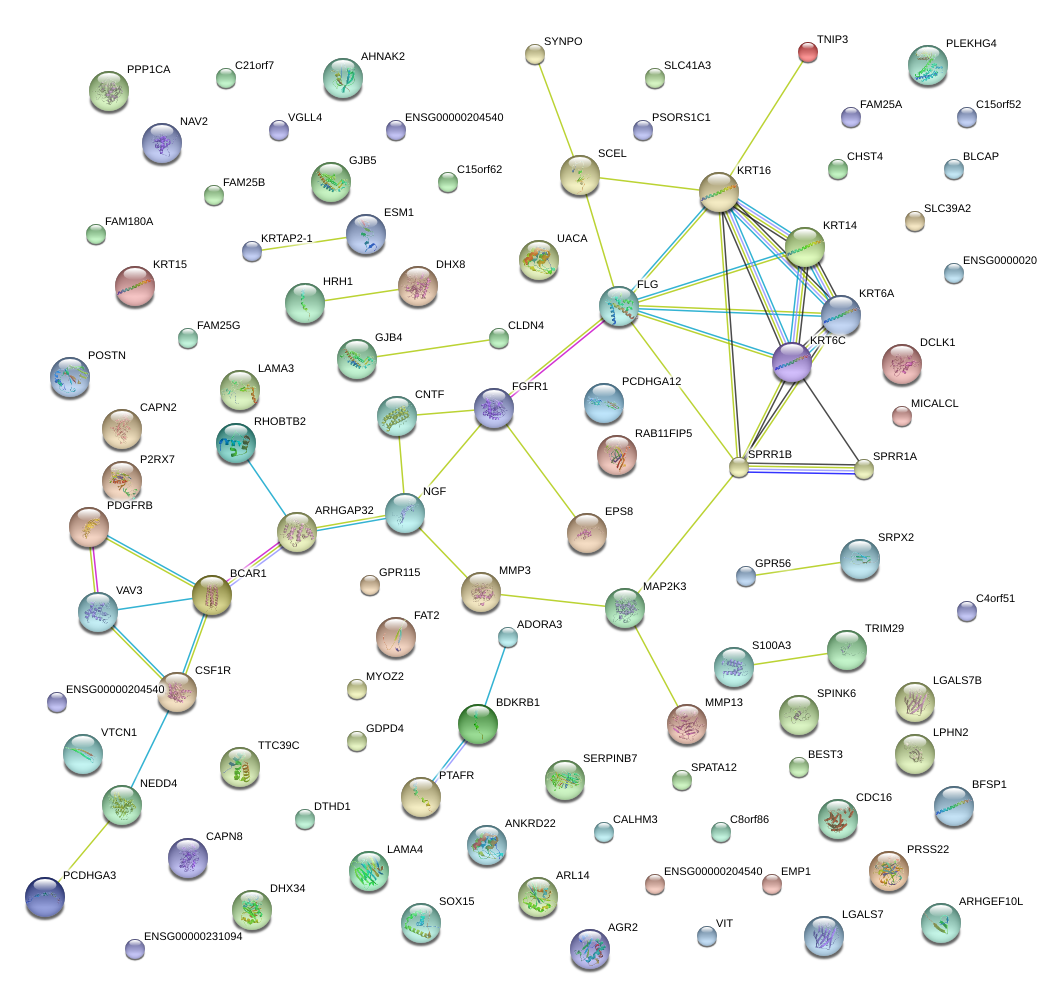
|
chr18_+_59593591
|
19.374
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr17_-_37022465
|
18.628
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr3_+_11153778
|
17.948
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr9_-_90456824
|
16.721
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr5_+_150000394
|
15.891
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr21_+_29439768
|
15.106
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr13_-_35327798
|
14.862
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr1_+_221955876
|
14.856
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr8_+_22900863
|
14.800
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr6_+_47774247
|
14.267
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr11_-_102219450
|
14.162
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr12_-_68369322
|
13.410
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr7_+_72883128
|
13.317
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr1_+_17719405
|
13.240
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr17_-_36469869
|
13.172
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr17_+_21131940
|
13.082
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr18_+_19947314
|
12.967
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr13_+_77007809
|
12.358
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr3_-_11585264
|
12.226
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr5_+_140848905
|
12.181
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr14_-_104507862
|
11.939
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr12_+_13240868
|
11.880
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr8_-_38505331
|
11.527
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr5_+_147562531
|
11.476
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr17_-_36996611
|
11.302
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr12_-_51173238
|
11.236
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr20_-_17487480
|
11.171
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr3_+_57069508
|
11.137
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr14_+_95792299
|
10.979
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr4_+_120276386
|
10.904
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr7_-_135083906
|
10.555
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr15_+_38849450
|
10.466
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr15_-_68781662
|
10.433
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr19_-_43955984
|
10.406
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr4_-_122304925
|
9.969
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr11_+_12265022
|
9.962
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr1_-_36589744
|
9.819
|
|
|
|
|
chr2_-_73170290
|
9.774
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr1_-_115682374
|
9.767
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr9_+_676640
|
9.751
|
|
|
|
|
chr15_-_53996597
|
9.545
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr1_-_151788341
|
9.342
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr19_-_40711034
|
9.302
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chrX_+_99786044
|
9.212
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_+_151270301
|
9.188
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr14_+_20537253
|
9.148
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr16_+_56219802
|
9.135
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr12_+_13240983
|
8.954
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_34993234
|
8.912
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr1_-_108032645
|
8.847
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr4_+_35959638
|
8.804
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr10_-_88720908
|
8.779
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr1_-_221920023
|
8.738
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chrX_+_99785818
|
8.737
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr10_-_105228986
|
8.614
|
NM_001129742
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr6_+_31190505
|
8.609
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr11_-_119499188
|
8.601
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr12_+_13240987
|
8.588
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_117554999
|
8.536
|
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr5_-_149473127
|
8.450
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr18_+_19706979
|
8.438
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr7_-_16811131
|
8.344
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr10_-_90601655
|
8.343
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr12_-_15706340
|
8.307
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr2_+_36777336
|
8.279
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr16_-_2848106
|
8.160
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr1_+_34997928
|
8.152
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr11_+_58146721
|
8.100
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr6_-_169392771
|
7.944
|
|
|
|
|
chr15_-_38417564
|
7.935
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr16_+_65868913
|
7.811
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr11_-_76676110
|
7.793
|
NM_182833
|
GDPD4
|
glycerophosphodiester phosphodiesterase domain containing 4
|
|
chr5_-_54317102
|
7.527
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr13_-_37070862
|
7.501
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr20_-_35586366
|
7.469
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr1_-_111908085
|
7.410
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr1_-_150564302
|
7.392
|
NM_002016
|
FLG
|
filaggrin
|
|
chr3_+_161877640
|
7.268
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr4_+_146820805
|
7.194
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr1_-_28392966
|
7.100
|
NM_001164722
NM_001164721
|
PTAFR
|
platelet-activating factor receptor
|
|
chr16_+_70117523
|
7.088
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr17_-_7434113
|
7.057
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr1_-_221919982
|
7.031
|
|
CAPN8
|
calpain 8
|
|
chr10_+_88770025
|
7.022
|
NM_001146157
|
FAM25A
|
family with sequence similarity 25, member A
|
|
chr11_-_128399218
|
6.959
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr17_-_36928604
|
6.828
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr3_-_127258299
|
6.805
|
NM_001008487
|
SLC41A3
|
solute carrier family 41, member 3
|
|
chr16_-_73859451
|
6.720
|
NM_001170715
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr1_+_81544432
|
6.630
|
|
LPHN2
|
latrophilin 2
|
|
chr11_-_102331643
|
6.629
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr11_+_20005704
|
6.617
|
|
NAV2
|
neuron navigator 2
|
|
chr10_+_24795446
|
6.615
|
|
KIAA1217
|
KIAA1217
|
|
chr9_-_34652629
|
6.577
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr10_+_24795465
|
6.536
|
|
KIAA1217
|
KIAA1217
|
|
chr15_+_65245546
|
6.512
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr11_-_101906640
|
6.496
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr19_-_56164628
|
6.439
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr2_-_202271598
|
6.422
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr1_+_151147646
|
6.275
|
NM_005547
|
IVL
|
involucrin
|
|
chr19_-_56204615
|
6.243
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr10_+_47867667
|
6.207
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr11_-_123353607
|
6.172
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr1_+_181421796
|
6.074
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr3_-_150578207
|
5.948
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr11_-_102174102
|
5.942
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr18_-_26996742
|
5.917
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr2_+_219180862
|
5.901
|
|
PLCD4
|
phospholipase C, delta 4
|
|
chr11_-_102174032
|
5.901
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr19_+_43971689
|
5.896
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr2_-_18634257
|
5.877
|
NM_001002006
NM_033253
|
NT5C1B-RDH14
NT5C1B
|
NT5C1B-RDH14 readthrough
5'-nucleotidase, cytosolic IB
|
|
chr3_-_188936941
|
5.836
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr14_+_51025604
|
5.813
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr15_+_38319326
|
5.796
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr19_-_50977485
|
5.792
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr1_+_40508336
|
5.752
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr1_+_181421984
|
5.720
|
|
LAMC2
|
laminin, gamma 2
|
|
chr6_+_41712907
|
5.712
|
|
MDFI
|
MyoD family inhibitor
|
|
chr18_-_45630154
|
5.705
|
|
MYO5B
|
myosin VB
|
|
chr7_+_1543644
|
5.669
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr3_-_48576204
|
5.563
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr22_-_41357380
|
5.518
|
|
|
|
|
chr4_-_152366970
|
5.495
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr8_-_72431274
|
5.386
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_+_47732181
|
5.374
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr18_+_50646616
|
5.350
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr3_+_185520744
|
5.282
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr2_+_202379442
|
5.271
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr11_-_8695974
|
5.260
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_+_151223180
|
5.206
|
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr20_+_19903778
|
5.187
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr18_+_59295123
|
5.178
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr4_+_661710
|
5.056
|
NM_002477
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr5_+_59819761
|
5.045
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr16_-_30983909
|
5.025
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr5_+_36644187
|
4.993
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_-_46601687
|
4.982
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr6_-_152744143
|
4.968
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr2_+_69055208
|
4.952
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr2_+_233605625
|
4.925
|
NM_005383
|
NEU2
|
sialidase 2 (cytosolic sialidase)
|
|
chr14_+_62740878
|
4.912
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr2_+_113591940
|
4.904
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr4_-_54119159
|
4.886
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr1_+_150753601
|
4.855
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr5_+_73145098
|
4.838
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr12_-_90097384
|
4.830
|
NM_133503
|
DCN
|
decorin
|
|
chr10_+_118177371
|
4.799
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr11_-_62213946
|
4.798
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr9_-_129381086
|
4.782
|
NM_001035534
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr4_-_151990061
|
4.779
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr3_+_190147633
|
4.759
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr21_+_25856311
|
4.748
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chrX_-_33139349
|
4.720
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_-_159325709
|
4.712
|
|
PVRL4
|
poliovirus receptor-related 4
|
|
chr4_+_76700281
|
4.700
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr5_+_35892747
|
4.678
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr4_-_100575313
|
4.644
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr12_+_106498763
|
4.635
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr16_-_30032330
|
4.620
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr19_-_50977592
|
4.613
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr2_+_166034402
|
4.611
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr9_-_90983501
|
4.603
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr6_+_108593907
|
4.598
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr17_-_53335748
|
4.587
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr2_+_169634330
|
4.538
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr5_-_59819642
|
4.520
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_185521139
|
4.509
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_105057940
|
4.504
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr22_-_43986896
|
4.449
|
|
KIAA0930
|
KIAA0930
|
|
chr15_+_39918302
|
4.440
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr9_+_35663852
|
4.433
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr3_-_150578148
|
4.432
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr6_+_30964441
|
4.421
|
NM_013994
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr3_-_197549640
|
4.389
|
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr19_-_50977586
|
4.387
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr19_-_60573552
|
4.335
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr3_+_101720753
|
4.327
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr20_+_34206070
|
4.321
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr3_-_197549588
|
4.309
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr8_-_143848225
|
4.232
|
NM_177458
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chrX_-_19598912
|
4.226
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_181422016
|
4.223
|
|
LAMC2
|
laminin, gamma 2
|
|
chr10_-_48877824
|
4.222
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr3_+_64645819
|
4.188
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr3_-_150578103
|
4.186
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr17_-_37034288
|
4.174
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr1_+_24518501
|
4.133
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr4_+_40032146
|
4.113
|
NM_017581
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9
|
|
chr11_-_62213577
|
4.111
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr9_-_116920232
|
4.110
|
NM_002160
|
TNC
|
tenascin C
|
|
chr10_-_116241572
|
4.088
|
|
|
|
|
chr1_+_151025376
|
4.062
|
NM_178353
|
LCE1E
|
late cornified envelope 1E
|
|
chr10_-_31328426
|
4.049
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr10_-_62002672
|
4.047
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_+_74462226
|
4.026
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr15_+_84021271
|
4.024
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr13_-_85271329
|
4.017
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr13_+_96592051
|
3.995
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_-_58175437
|
3.960
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr1_+_24518398
|
3.932
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr19_-_48661563
|
3.927
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|